38 r plot tree with labels
Label orientation in an R data.tree plot - Stack Overflow library ("data.tree") data (acme) setgraphstyle (acme, rankdir = "tb") setedgestyle (acme, arrowhead = "vee", color = "blue", penwidth = 2) #per default, node style attributes will be inherited: setnodestyle (acme, style = "filled,rounded", shape = "box", fillcolor = "greenyellow", fontname = "helvetica", tooltip = getdefaulttooltip) setnodestyle … plot.phylo function - RDocumentation (= labels for unrooted trees) a character string specifying the display of tip labels for unrooted trees (can be abbreviated): either "horizontal" where all labels are horizontal (the default if type = "u" ), or "axial" where the labels are displayed in the axis of the corresponding terminal branches.
nodelabels: Labelling the Nodes, Tips, and Edges of a Tree ... R Documentation Labelling the Nodes, Tips, and Edges of a Tree Description These functions add labels to or near the nodes, the tips, or the edges of a tree using text or plotting symbols. The text can be framed. Usage
R plot tree with labels
R Decision Trees Tutorial: Examples & Code in R for ... Step 3: Lastly, you use an average value to combine the predictions of all the classifiers, depending on the problem. Generally, these combined values are more robust than a single model. While bagging can improve predictions for many regression and classification methods, it is particularly useful for decision trees. Phylogenetic trees in R - Sensory Evolution This matrix represents unique combinations of node- and tip numbers, defining each branch segment of the tree. plot (mini.phy, label.offset=0.2) # the label.offset argument moves the names a bit to the right nodelabels () # add node numbers tiplabels () # add tip numbers Treemap charts in R - Plotly There are three different attributes you can use to change the color of the sectors of treemaps you have created with Plotly for R: marker.colors marker.colorscale colorway, The following examples show how to use each attribute. To use marker.colors, pass a list of valid CSS colors or hexadecimal color codes.
R plot tree with labels. Changing phylogeny tip labels in R - R-bloggers The following is a simple R -based solution for changing the tip labels of phylogenetic trees. First, we need to create a tree and a dataframe containing both the specimen codes and the ultimate labels. library (ape) tr <- rtree (5) d1 <- c ("t1","t2","t3","t4","t5") nodelabels function - RDocumentation nodelabels: Labelling the Nodes, Tips, and Edges of a Tree Description These functions add labels to or near the nodes, the tips, or the edges of a tree using text or plotting symbols. The text can be framed. Usage PDF Plotting rpart treeswiththe rpart.plot package 6 Customizing the node labels 13 7 Examples using the color and palette arguments 18 8 Branch widths 27 9 Trimming a tree with the mouse 28 10Using plotmoin conjunction with prp 29 11Compatibility with plot.rpartand text.rpart 32 12The graph layout algorithm 33 An Example temp < 68 ibh >= 3574 dpg < −9 ibt < 227 temp >= 68 ibh < 3574 dpg ... Powerful tree graphics with ggplot2 - GitHub Pages Tip labels. label.tips - The label.tips parameter controls labeling of tree tips (AKA leaves). Default is NULL, indicating that no tip labels will be printed. If "taxa_names" is a special argument resulting in the OTU name (try taxa_names function) being labelled next to the leaves or next to the set of points that label the leaves. Alternatively, if your data object contains a tax_table, then ...
Visualizing and Annotating Phylogenetic Trees with R+ggtree The ggtree Package. ggtree is an R package that extends ggplot2 for visualizating and annotating phylogenetic trees with their covariates and other associated data. It is available from Bioconductor.Bioconductor is a project to provide tools for analyzing and annotating various kinds of genomic data. You can search and browse Bioconductor packages here. R: Plot a taxonomic tree Tree labels are displayed over their tree. Accepts a vector, the same length taxon_id or a factor of its length. sizes The size of nodes, edges, labels, and trees can be mapped to various conditions. This is useful for displaying statistics for taxa, such as abundance. Only the relative size of the condition is used, not the values themselves. R plot() Function (Add Titles, Labels, Change Colors and ... Adding Titles and Labeling Axes We can add a title to our plot with the parameter main. Similarly, xlab and ylab can be used to label the x-axis and y-axis respectively. plot (x, sin (x), main="The Sine Function", ylab="sin (x)") Changing Color and Plot Type We can see above that the plot is of circular points and black in color. plot - R: plotting decision tree labels leaves text cut ... Basically, you have to mutate age and fare columns into numeric variables. Like: clean_titanic <- titanic %>% select (-c (home.dest, cabin, name, x, ticket)) %>% mutate ( pclass = factor (pclass, levels = c (1, 2, 3), labels = c ('Upper', 'Middle', 'Lower')), survived = factor (survived, levels = c (0, 1), labels = c ('No', 'Yes')), # HERE.
How to label specific points in scatter plot in R ... The plot() method in Base R is used to plotting the R objects, namely, lists or data frames. Syntax: plot(x, y, data , col) Parameter : x,y - The x and y coordinates of the points. col - The color to assign to the points. The color is specified using a character string. data - The data frame points to be plotted in the graph R: Plot a tree with colored internal node labels using ggtree R: Plot a tree with colored internal node labels using ggtree plotTrees {dowser} R Documentation Plot a tree with colored internal node labels using ggtree Description plotTrees plots a tree or group of trees Usage plot.tree function - RDocumentation Plot a tree object on the current graphical device Usage # S3 method for tree plot (x, y = NULL, type = c ("proportional", "uniform"), ...) Arguments x an object of class "tree". y ignored. Used for positional matching of type. type character string. If this partially matches "uniform", the branches are of uniform length. plot_tree function - RDocumentation plot_tree (physeq, method = "sampledodge", nodelabf = NULL, color = NULL, shape = NULL, size = NULL, min.abundance = Inf, label.tips = NULL, text.size = NULL, sizebase = 5, base.spacing = 0.02, ladderize = FALSE, plot.margin = 0.2, title = NULL, treetheme = NULL, justify = "jagged") Arguments physeq (Required).
text.tree function - RDocumentation Description Add text to a tree plot. Usage # S3 method for tree text (x, splits = TRUE, label = "yval", all = FALSE, pretty = NULL, digits = getOption ("digits") - 3, adj = par ("adj"), xpd = TRUE, ...) Arguments x an object of class "tree" splits logical. If TRUE the splits are labelled label
Draw Scatterplot with Labels in R (3 Examples) | Base R ... In this post, I'll explain how to add labels to a plot in the R programming language. The article consists of three examples for the addition of point labels. To be more precise, the table of content looks like this: 1) Creating Example Data. 2) Example 1: Add Labels to Base R Scatterplot. 3) Example 2: Add Labels to ggplot2 Scatterplot.
plot.phylo: Plot Phylogenies in ape: Analyses of ... The font format of the labels of the nodes and the tips is the same. If no.margin = TRUE, the margins are set to zero and are not restored after plotting the tree, so that the user can access the coordinates system of the plot. The option 'node.pos' allows the user to alter the vertical position (i.e., ordinates) of the nodes.
PLOT in R ⭕ [type, color, axis, pch, title, font, lines ... R plot x and y labels. By default, R will use the vector names of your plot as X and Y axes labels. However, you can change them with the xlab and ylab arguments. plot(x, y, xlab = "My X label", ylab = "My Y label") If you want to delete the axes labels you can set them to a blank string or set the ann argument to FALSE.
katholt/plotTree - GitHub Tree options (see ?plot.phylo in R for more info) • tip.labels = T turns on printing strain names at the tips • tipLabelSize = 1 change the size of printed strain names (only relevant if tip.labels=T) • offset=0 change the spacing between the end of the tip and the printed strain name (only relevant if tip.labels=T)
Chapter 7 Plotting tree with data | Data Integration ... 7.2 Aligning Graph to the Tree Based on the Tree Structure. For associating phylogenetic tree with different types of plot produced by user's data, ggtree provides geom_facet() layer and facet_plot() function which accept an input data.frame and a geom layer to draw the input data. The data will be displayed in an additional panel of the plot.
Drawing a tree with colored tips in R ... - Anita Schürch ggtree is a R package designed for viewing and annotating phylogenetic trees. It is based on the ggplot2 package. Here I will show step-by-step how to draw a tree with colored tips, because, even thought the bioconductor documentation is clear and complete, I did not read about this very common visualization.
text.tree: Annotate a Tree Plot in tree: Classification ... Add text to a tree plot. Usage 1 2 3 4 ## S3 method for class 'tree' text (x, splits = TRUE, label = "yval", all = FALSE, pretty = NULL, digits = getOption ("digits") - 3, adj = par ("adj"), xpd = TRUE, ...) Arguments Details If pretty = 0 then the level names of a factor split attributes are used unchanged.
R: Plot Clustering Tree of a Hierarchical Clustering Details. Creates a plot of a clustering tree given a twins object. The leaves of the tree are the original observations. In case of an agglomerative clustering, two branches come together at the distance between the two clusters being merged. For a divisive clustering, a branch splits up at the diameter of the cluster being splitted.
Treemap charts in R - Plotly There are three different attributes you can use to change the color of the sectors of treemaps you have created with Plotly for R: marker.colors marker.colorscale colorway, The following examples show how to use each attribute. To use marker.colors, pass a list of valid CSS colors or hexadecimal color codes.
Phylogenetic trees in R - Sensory Evolution This matrix represents unique combinations of node- and tip numbers, defining each branch segment of the tree. plot (mini.phy, label.offset=0.2) # the label.offset argument moves the names a bit to the right nodelabels () # add node numbers tiplabels () # add tip numbers
R Decision Trees Tutorial: Examples & Code in R for ... Step 3: Lastly, you use an average value to combine the predictions of all the classifiers, depending on the problem. Generally, these combined values are more robust than a single model. While bagging can improve predictions for many regression and classification methods, it is particularly useful for decision trees.








![[PYTHON] Visualize scikit-learn decision trees with Plotly's Treemap](https://qiita-image-store.s3.ap-northeast-1.amazonaws.com/0/218449/fe43cf2b-a2c3-65bc-15b4-67502c8112d4.png)


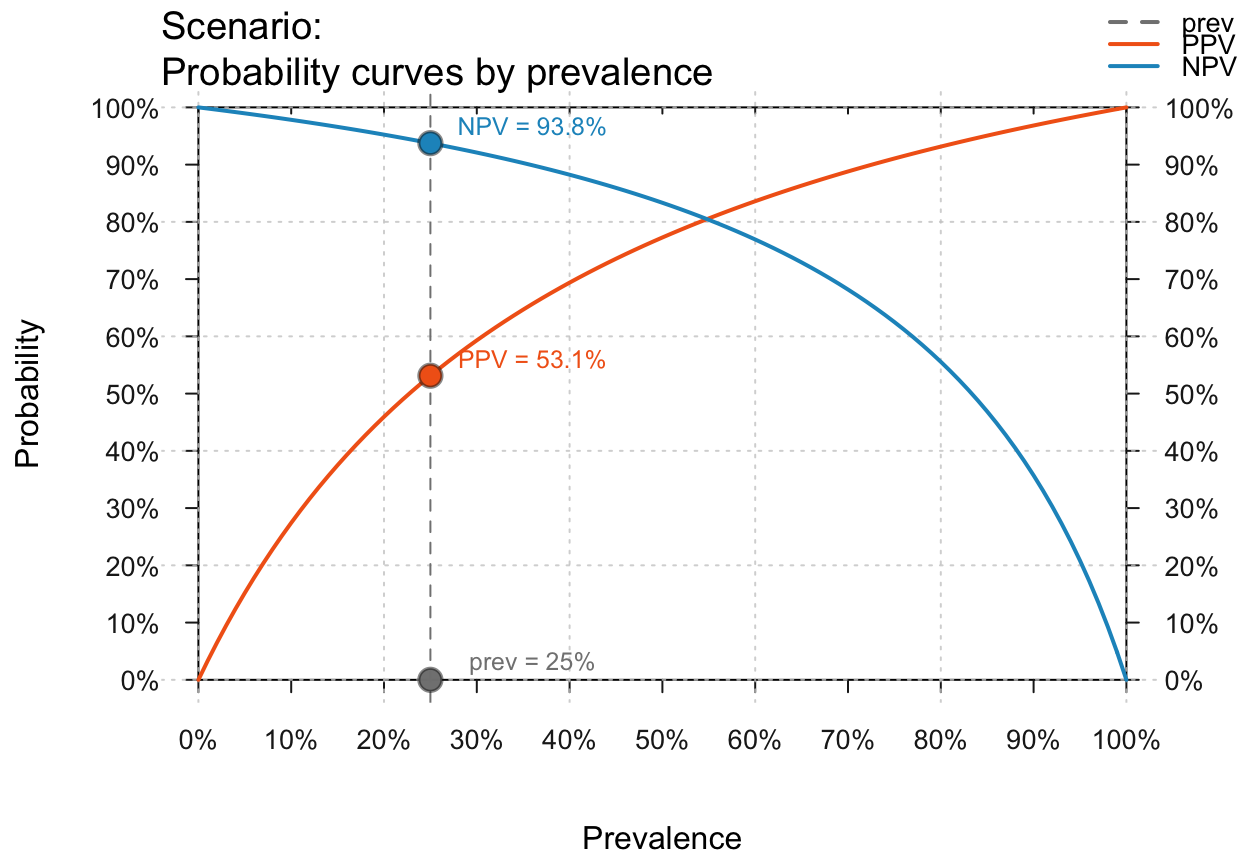
Post a Comment for "38 r plot tree with labels"